FastSemSim API¶
The layer of “entrypoint functions” (API) available in FastSemSim conveniently allows interacting with ontologies, Annotation Corpora (ACs) and semantic similarity without knowing the inner workings of the FastSemSim library.
This section describes these entrypoint functions and explains how to use them. Examples and use cases are presented as well.
The standard processing workflow used when calculating semantic similarity scores can be recapitulated in the following 6 steps:
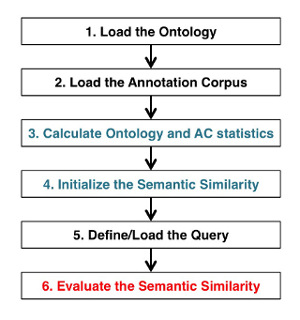
For each of these points, some entrypoint functions take care of masking all the inner workings of the package. It is noteworthy to note that albeit the primary function of FastSemSim is calculating semantic similarity, its representation of ontologies and ACs can be used as a base to extract statistics. explore the data or perform other analyses based on ontology annotations.
0. Importing FastSemSim¶
After installing the pacakge, you can import FastSemSim in your Python environment with:
import fastsemsim
1. API - Ontologies¶
Loading an ontology¶
Loading an ontology from a file or a descriptor (more about descriptors in section FastSemSim Datasets) is as simple as running a single line of code using the entrypoint function fastsemsim.load_ontology:
my_ontology = fastsemsim.load_ontology(...)
If invoked without parameters, this function will load the Gene Ontology shipped with fastsemsim. Passing the right parameters to the function allows loading other ontologies included in fastsemsim, as well as custom ontologies.
Here the full description of the function:
2. API - Annotation Corpora¶
Loading an annotation corpus¶
Loading an annotation corpus (AC) is as easy as loading ontologies. The only additional requirement is that an ontology must be passed as parameter to the parser, that will take care of matching the terms in the ontology to the terms in the annotation corpus. The AC can be loaded from a file or from a descriptor (more about descriptors in section FastSemSim Datasets) using the entrypoint function fastsemsim.load_ac:
ac = fastsemsim.load_ac(ontology = my_ontology, ...)
If invoked without parameters, beside the mandatory ontology parameter, this function will load one of the ACs shipped with fastsemsim. Passing the right parameters to the function allows loading other ontologies included in fastsemsim (see Dataset section), as well as custom ontologies from a file. In general, fastsemsim is able to load any file coming from the geneontology community (gaf2 file format).
Here the full description of the function:
3. Data embedded in FastSemSim¶
Fastsemsim includes some of the standard broadly used ontologies and annotation corpora. The corresponding files are automatically installed with the library.
All the available ACs and ontologies are managed by the Dataset class. The Dataset class works around the concept of Descriptor. Each ontology or AC available is referenced with a Descriptor indicating where the file is stored and the parameters necesary to correctly load it. Upon importing fastsemsim, an instance of the class Dataset is automatically created and filled with the descriptors of the Acs and ontologies embedded in fatsemsim. Such Dataset object is available as:
fastsemsim.dataset
The Dataset class exposes some convenient functions to easily interrogate the available ACs and ontologies and load them. It is possible, for instance, to list the available ontologies of a given type (e.g. GeneOntology, DiseaseOntology, …) and get a valid descriptor. The descriptor can be passed to the ontology or AC loading functions (see API - Ontoliges and API - Annotation Corpora sections).
The main functions exposed by Dataset are:
- list_ontologies(): list the ontologies available in fastsemsim.
fastsemsim.dataset.list_ontologies()
- list_acs(): list the ACs available in fastsemsim, you can use the function .
fastsemsim.dataset.list_acs()
- get_ontology(): returns the descriptor(s) of the ontologies satisfying the passed parameters (if any).
Note that more than one descriptor might be returned.
fastsemsim.dataset.get_ontology(ontology_type=...)
fastsemsim.dataset.get_ontology(ontology_type='GeneOntology') # to look specifically for the Gene Ontology
- get_annotation_corpus(): returns the descriptor(s) of the annotation corpora satisfying the passed parameters (if any).
Note that more than one descriptor might be returned.
fastsemsim.dataset.get_annotation_corpus(ontology_type=..., ac_species=...)
fastsemsim.dataset.get_annotation_corpus(ontology_type='GeneOntology', ac_species='human') # look for ACs compatible with the Gene Ontology, for the human species
Here the full description of the four functions list above:
For more details about the Dataset class, check the full documentation of the data module available at the follwing page: